Portfolio
Project Overview:
Overview:
Recognizing emotions from voice alone is a complex task, given the subjective and non-universal nature of emotions. This project tackles this challenge by building a model to accurately classify the emotional content of vocal expressions, despite the absence of facial expressions and body language cues.
Datasets:
The project utilizes four diverse datasets: RAVDESS, TESS, SAVEE, and CREMA-D. This diversity enhances the model’s ability to generalize across different emotional expressions.
Models:
Three models are employed to classify emotions:
- A Convolutional Neural Network (CNN) trained on Mel Spectrograms.
- A CNN trained on the Mel Frequency Cepstral Coefficients (MFCCs).
- A Convolutional Recurrent Neural Network (CRNN) trained on MFCCs.
Findings:
The Mel Spectrogram CNN model demonstrated good performance but faced challenges in distinguishing certain emotions.
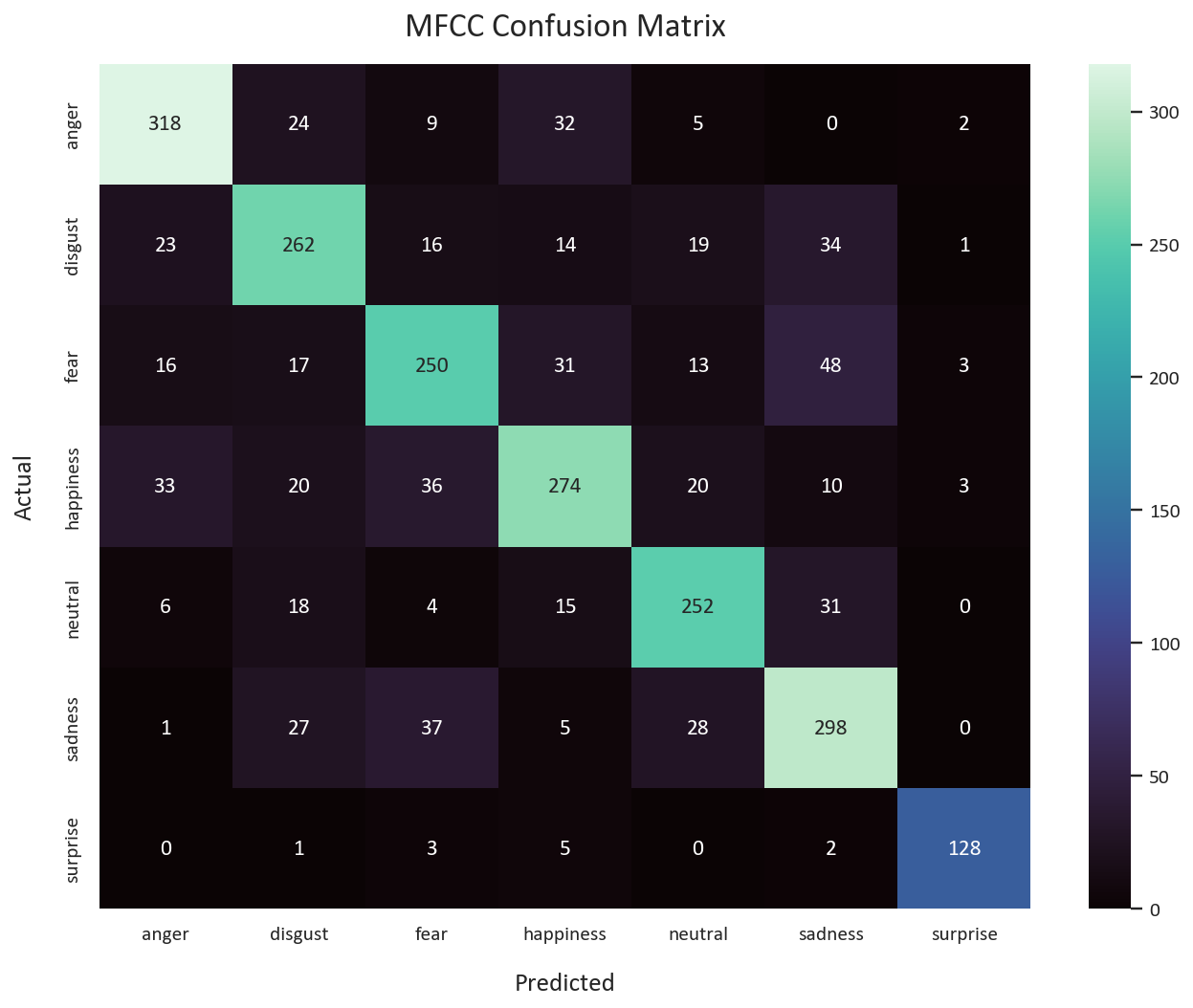
The MFCCs CNN model outperformed the others, with a performance close to state-of-the-art. indicating the effectiveness of MFCCs as a representation for this task.
The MFCCs CRNN model performed well but did not surpass the MFCCs CNN model and tended to overfit
Evaluation:
Precision, Recall, and F1 score metrics were used for model evaluation:
- Mel Spectrogram Model:
- Accuracy: 71%
- Macro Average F1-score: 0.73
- MFCCs CNN Model:
- Accuracy: 74%
- Macro Average F1-score: 0.76
- MFCCs CRNN Model:
- Accuracy: 74%
- Macro Average F1-score: 0.75
Summary: The project successfully tackles the complexities of speech-based emotion recognition, emphasizing the effectiveness of training CNNs on MFCCs. The best model achieves performance close to state of the art.
The entire project can be found in this github repository:
Project Overview:
Overview:
In this project, the primary goal is to use Convolutional Neural Networks (CNNs), for the detection of thoracic diseases through the analysis of chest X-ray images. The inherent challenge lies in handling imbalanced datasets, where the number of normal cases significantly outweighs that of disease cases. This imbalance poses a critical issue as a seemingly accurate model could, in reality, exhibit poor performance in predicting diseases, risking misdiagnosis and potentially fatal consequences, especially in the medical domain.
Dataset:
The project utilizes the extensive NIH Chest X-ray dataset, comprising 112,120 images, each labeled with up to 14 different diseases. These labels are derived through advanced Natural Language Processing (NLP) techniques, ensuring an expected accuracy of over 90%.
Disease Classes:
The model is designed to predict 14 distinct diseases, including conditions such as Atelectasis, Consolidation, Infiltration, Pneumothorax, Edema, Emphysema, Fibrosis, Effusion, Pneumonia, Pleural Thickening, Cardiomegaly, Nodule, Hernia, and Mass. Notably, the dataset analysis reveals varying percentages of each disease and sheds light on the dataset’s inherent imbalance, a crucial factor for subsequent model evaluations.
Models:
The project employs two distinct CNN models. The first utilizes binary cross-entropy loss, while the second is trained with weighted binary cross-entropy loss. These models are chosen for their potential in highlighting the challenges posed by imbalanced datasets.
- Binary Cross-Entropy Loss Model:
- Performance Insights: Despite achieving an impressive overall accuracy, this model exhibits a notable drawback in the medical context. While correctly identifying 98.54% of negative cases, its ability to pinpoint positive cases is concerning, with a mere 10.31% success rate. This elevated rate of false negatives raises alarms, as it could potentially lead to delayed treatment for genuinely ill patients.
- Weighted Binary Cross-Entropy Loss Model:
- Performance Insights: The weighted model strikes a better balance, correctly identifying 86.62% of positive cases. Although its overall accuracy is lower, identifying 44.84% of negative cases, the higher recall renders it more suitable for medical applications. The emphasis here is on reducing false negatives, even at the expense of an increased false positive rate.
Evaluation Metrics:
Given the dataset’s significant imbalance, we used different metrics:
- AUC ROC (Area Under the Receiver Operating Characteristic curve): An indicator of a model’s ability to discriminate between positive and negative cases.
- Precision: Focused on minimizing false positives, preventing false disease diagnoses.
- Recall: Emphasizes the ability to capture all positive cases, mitigating the risk of false negatives and subsequent delayed treatment.
Findings:
Evaluation of the models reveals crucial insights. The binary cross-entropy loss model, while achieving high overall accuracy, falters in identifying positive cases, with a mere 10.31% success rate. This high rate of false negatives raises significant concerns in a medical context, where timely treatment is paramount.
In contrast, the weighted binary cross-entropy loss model outperforms its counterpart, correctly identifying 86.62% of positive cases with a higher recall. Despite a dip in overall accuracy, the heightened sensitivity makes it more suitable for medical applications, prioritizing the reduction of false negatives, even at the cost of increased false positives.
Summary:
This project underscores the delicate balance between accuracy and sensitivity in the context of medical machine learning, with a clear emphasis on minimizing the risks associated with false negatives, which are generally more dangerous than false positives.
The entire project can be found in this github repository:
Project Overview:
Overview:
This project revolves around Sales Data Analysis and Forecasting, employing Ensemble Methods to predict future sales based on historical data. The project uses a dataset of 1,115 drugstores available on Kaggle.
Models:
- Random Forest Regressor: Robust ensemble method, averaging predictions from multiple decision trees to prevent overfitting.
- XGBRegressor: eXtreme Gradient Boosting Regressor, an iterative model known for its effectiveness in structured datasets.
Evaluation Metrics:
- Mean Absolute Error (MAE): Indicates the average absolute difference between predicted and actual values.
- Mean Absolute Percentage Error (MAPE): Expresses the average absolute error as a percentage of actual values.
- Symmetric Mean Absolute Percentage Error (SMAPE): A symmetric variation of MAPE addressing its limitations.
- R2 Score (Coefficient of Determination): Measures the proportion of predictable variance in the dependent variable.
Findings:
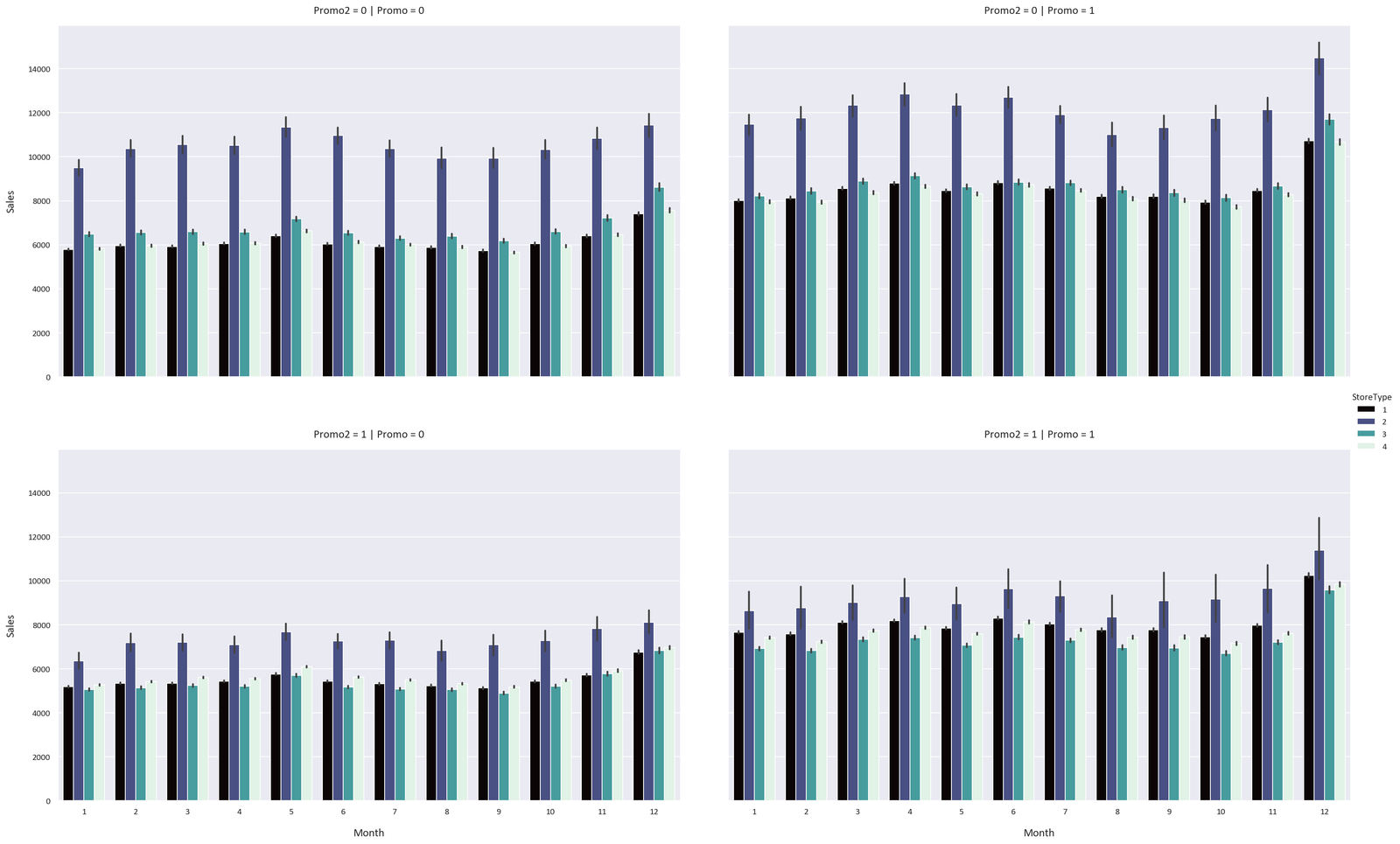
Both models perform extremely well, with the XGBoost regressor performing slightly better. This is because the XGBoost regressor is an iterative model, so it can learn from its mistakes and improve its predictions. It learnt that the Promo and Promo2 features are very important, while the random forest regressor didn’t learn this.
- The MAE of the random forest regressor is 1115, with a SMAPE of 16.12% and an R2 score of 0.70.
- The MAE of the XGBoost regressor is 1031, with a SMAPE of 15.12% and an R2 score of 0.73.
Summary:
This project showcases the power of ensemble methods in sales forecasting while providing valuable insights into customer behavior, seasonal patterns, and the impact of promotional strategies.
The entire project can be found in this github repository: